Overview
Overview
simBio is a tool for making it easier to build cell organism models, and for promoting the sharing/reuse of models. It provides the necessary features to create models which use ordinary differential equations and mathematical models, do integral calculation, display result graphs, extract results to files, change parameters and do calculations.
The structure of simBio is divided into 3 main elements. The first describes calculations of mathematical models with Java. The second describes the initial values of parameters and the structure of mathematical models with XML files. The third solves initial value problems of ordinary differential equations with an integration engine.
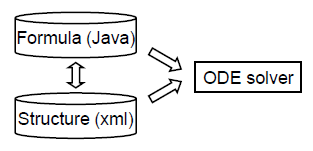
A mathematical model is created and run in 2 steps.
Java Files
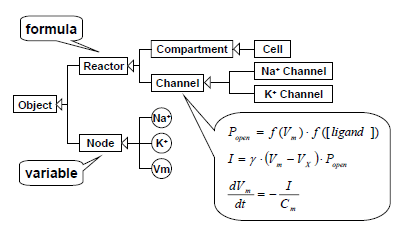
When a mathematical model is created, first Java is used to make a mathematical model of each function element based on biological knowledge.
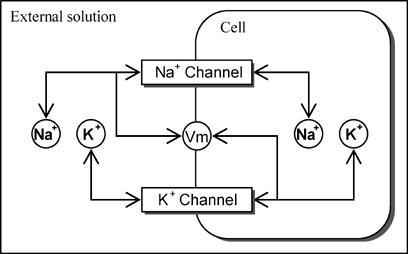
Using object-oriented principles, a function element model is described below Reactor, and a biological hierarchy is arranged in a modelled format. The figure displays a differential equation with the Channel element described. A formula specific to each channel is described in particular function elements. Then integral variables are defined as Nodes.
XML Files
Next, the initial values and interrelationships of function elements and variables are described using XML. Arrows in the figure show the interrelation. Function elements are freely combined to create a mathematical model, and run it.