news
simBio 1.0PR2 released
A new release of simBio is now available at http://sourceforge.net/project/showfiles.php?group_id=146713
Changes from previous version are
- The volume regulation model by Takeuchi et al (2006) was included. See instructions for usage.
- The calcium gate model of Gap junction by Oka et al (2006) was included.
- The analysis of NCX knockout by Sarai et al 2006 was included.
- The human ventricular cell models proposed by Kurata et al (2005) and by ten Tusscher et al (2004) were included.
- The Gap Junction model by Henriquez AP et al (2001) was included.
- The Japanese documents were separated to docs_ja project.
- The Documents were revised.
For more information, see http://sourceforge.net/projects/simbio/
Please stay tuned
The volume regulation model has been published in Takeuchi et al. (2006) on 1st Nov.
We are working hard to catch up The Journal of General Physiology, and the source code to reproduce the figures will be available in a few weeks.
Ayako Takeuchi, Shuji Tatsumi, Nobuaki Sarai, Keisuke Terashima, Satoshi Matsuoka, and Akinori Noma,
Ionic Mechanisms of Cardiac Cell Swelling Induced by Blocking Na+/K+ Pump As Revealed by Experiments and Simulation.
The Journal of General Physiology, 128(5): 495-507, 2006. doi:10.1085/jgp.200609646
simBio 0.3.02 released.
A new release of simBio is now available at http://sourceforge.net/project/showfiles.php?group_id=146713
Changes from previous version are
- The human cardiac cell model proposed by ten Tusscher et al (2004) is included at src/xml/tenTusscher_et_al_2004, see http://www.sim-bio.org/docs/bio/ for other models.
- Be able to modify a model xml to change settings from control. see src/xml/matsuoka_et_al2003/Fig.2.make.xml, also src/xml/kuratomi_et_al_2003.xml is deleted and the modification from matsuoka_et_al_2003 is written in kuratomi_et_al_2003.make.xml
- Add some tests.
- A duplicated jar is deleted.
- Some Javadoc comments are revised.
- Bugs are fixed, such as Ist, Ito, GUI.
For more information, see http://sourceforge.net/projects/simbio/
Eclipse Plug-in for simBio
Description
Execute simBio through pop-up menu of Eclipse Java Perspective. RunGUI, Composer, ResultGenerator, and XmlGenerator can be launched.
You can find latest information from http://www.sim-bio.org/.
License
This plugin is free software; you can redistribute it and/or modify it under the terms of the GNU Lesser General Public License as published by the Free Software Foundation; either version 2.1.
This plugin is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License (LGPL_2-1.txt) for more details.
The GNU Lesser General Public License can be found at http://www.gnu.org/copyleft/lesser.html
Requirements
- Eclipse 3.x (this plugin has created on Eclipse 3.0.2.)
- simBio (this plugin has created with simBio 0.2.2.)
Install
This plugin can be installed through update manager of Eclipse 3.x. Here the process with Eclipse 3.1 is described.
- Select Eclipse menu: "Help" - "Software Updates" - "Find and Install"
- Check "Search for new features to install", then "Next"
- Click "New Remote Site"
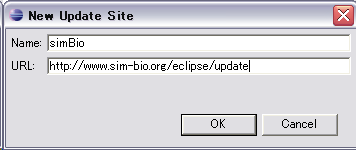 Input "Name:" box as you like such as "simBio",
and "URL:" box as "http://www.sim-bio.org/eclipse/update/", then "Finish"
Input "Name:" box as you like such as "simBio",
and "URL:" box as "http://www.sim-bio.org/eclipse/update/", then "Finish"
- "Updates" dialog will appear, so check simBio, then "Next".
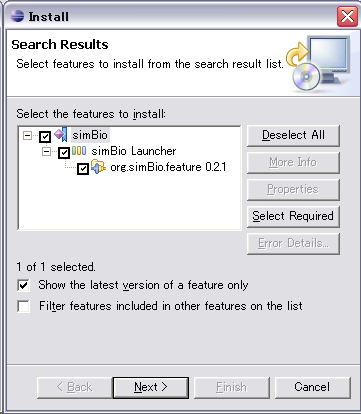
- Check "I accept the terms in the license agreement", then "Next"
- Click "Finish"
- "Verification" dialog will appear as not yet digitally signed.
- Click "Install All".
- "Install/Update" dialog will appear, You can choose either "Yes" or "Apply Changes".
Unpack simBio_eclipse_x.x.x.zip and put org.simBio.eclipse_* directory in Eclipse/plugins directory.
Usage
Run on GUI
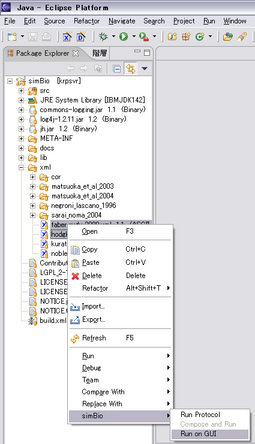 Right click xml files for simBio, then pop-up menu will appear.
Select [simBio] - [Run on GUI], then the XMLs are executed on GUI.
Right click xml files for simBio, then pop-up menu will appear.
Select [simBio] - [Run on GUI], then the XMLs are executed on GUI.
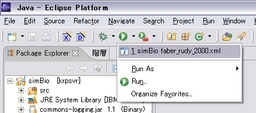 The Setting will be stored, then you can run it through the cool bottun.
The Setting will be stored, then you can run it through the cool bottun.
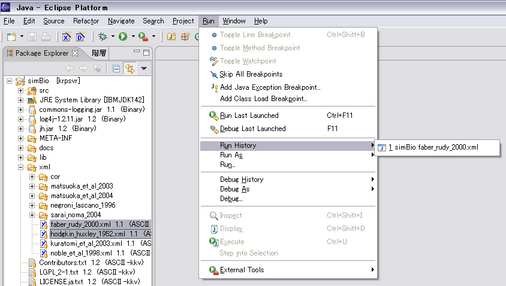 You can also run the previous settings through the menu, Run - Run History.
You can also run the previous settings through the menu, Run - Run History.
Compose and Run
Right click an xml file for org.simBio.Composer, select [simBio] - [Compose and Run], then a model xml will be composed and be run on GUI. An "composed.xml" file will be created on the same directory, so you need to reflesh the view of Eclipse.
Run Protocol
Right click an model xml and an protocol xml, select [simBio] - [Run Protocol], then org.simBio.ResultGenerator will run the protocol.
Generate XML
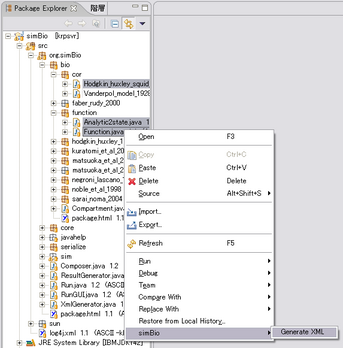 Right click Reactors and/or Analyzers,
select [simBio] - [Generate XML], then XML files for the specific class will be created.
Right click Reactors and/or Analyzers,
select [simBio] - [Generate XML], then XML files for the specific class will be created.
Copyright
Copyright (C) 2002-2005 Nobuaki Sarai and Contributors. The specific names are written in each file.
API
Javadoc API is located at http://www.sim-bio.org/docs/api/.
LRd and Noble98 models
The guinea-pig ventricular cell models of Luo-Rudy and Noble98 are incorpolated.
Faber GM, Rudy Y.
Action potential and contractility changes in [Na+]i overloaded cardiac myocytes: a simulation study.
Biophy J 2000;78:2392-2404
Noble D, Varghese A, Kohl P, Noble P.
Improved guinea-pig ventricular cell model incorporating a diadic space, IKr and IKs, and length- and tension-dependent processes.
Can J Cardiol. 1998 Jan;14(1):123-34.