download
simBio 0.3 released
simBio is a simulator for biological systems such as cardiac cells, epithelial cells, and pancreatic beta cells. simBio is written in Java, uses XML and can solve ordinary differential equations.
Changes from previous version are
- LGPL is applied.
- GUI has been improved.
- The volume regulation model of the ventricular myocyte (Terashima et al, 2006, in press) is included.
- Comments in xml files will be kept after calculation.
Available model list: http://www.sim-bio.org/docs/bio/
Download: http://sourceforge.net/projects/simbio/
Users' mailing list: http://sourceforge.net/projects/simbio/
In an attempt to mimic biological functional structures, a cell model is, in simBio, composed of independent functional modules called Reactors, such as ion channels and the sarcoplasmic reticulum, and dynamic variables called Nodes, such as ion concentrations. The interactions between Reactors and Nodes are described by the graph theory and the resulting graph represents a blueprint of an intricate cellular system. Reactors are prepared in a hierarchical order, in analogy to the biological classification. Each Reactor can be composed or improved independently, and can easily be reused for different models.
For more information, see
Nobuaki Sarai, Satoshi Matsuoka and Akinori Noma
simBio: a Java package for the development of detailed cell models
Progress in Biophysics and Molecular Biology, 2005, in press
0.2.1
Dr. Alan Garny has made a function that can translate a model in CellML into the simBio style, on Cellular Open Resource (COR).
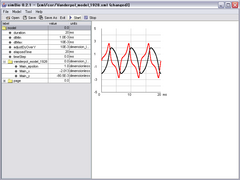
Screen shots
xml/cor/hodgkin_huxley_squid_axon_1952_modified.xml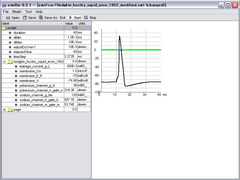
xml/cor/vanderpol_model_1928.xml
We made the latest version freely available.
Download: File List at SourceForge.net
Javadoc API has also updated.
The matsuoka_et_al_2004 model has slightly updated.
Matsuoka et al., 2004
The excitation–contraction–metabolism coupling model from Matsuoka et al., 2004 is incorpolated.
Download: File List at SourceForge.net
Matsuoka S, Sarai N, Jo H, Noma A.
Simulation of ATP metabolism in cardiac excitation-contraction coupling.
Prog Biophys Mol Biol. 2004 Jun-Jul;85(2-3):279-99.
- download simBio_*.zip and extract all.
- execute the simBio_*.jar included. (Java 2 Platform, Standard Edition, v 1.4.2 (J2SE) or later is needed, which is freely available from http://java.sun.com/.)
- select [File]-[Open] and open xml/matsuoka_et_al_2004/exp/AnoxiaSimulation.xml.
- select [Model]-[Start] to simulate anoxia.
Matsuoka et al, 2003
We are proud of making simBio available.
Download: File List at SourceForge.net
This package can reproduce the results in Matsuoka et al, 2003, which is a guinea pig ventricular cell model, and Sarai et al, 2003, which is a sino-atrial node cell model.
Please see docs folder included in the extracted files, for further information.